This demo consists of several Matlab scripts demonstrating how to download human microarray expression values from the API and identify the Brodmann Areas associated with the samples.
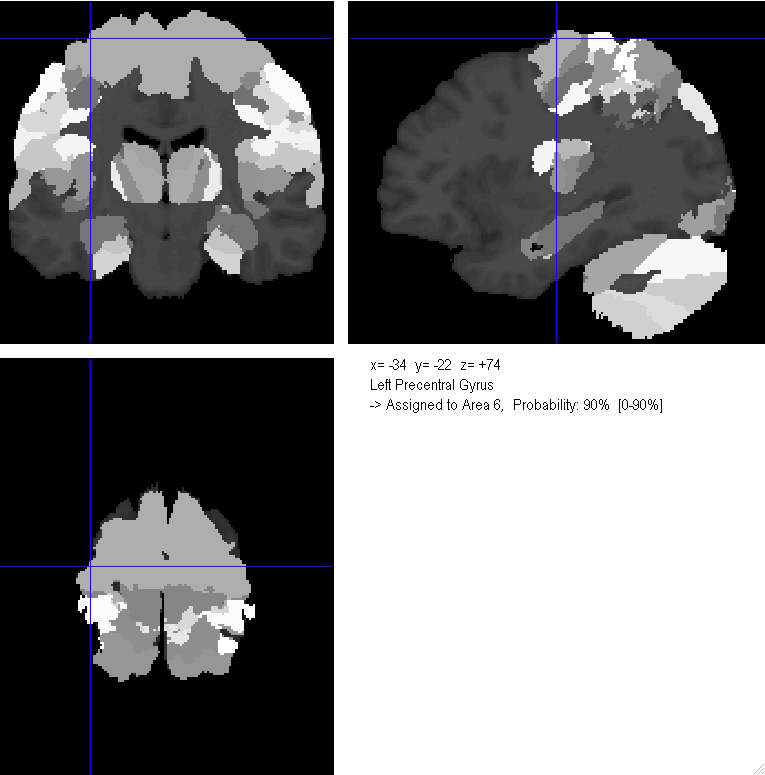
The Statistical Parametric Mapping (SPM) Matlab package is commonly used by the function brain imaging community for analysis. An additional Anatomy Toolbox adds several features to SPM, including probabilistic cytoarchitectonic area maps, which divide the cerebral cortex according to structural and cellular organization. For details on the toolbox and cytoarchitectonic maps, see the following references: (1) (2).
The Matlab scripts in this demo perform two API queries: one to download human specimen MNI transforms, and another to download microarray sample locations. Those locations are transformed to MNI and exported in a text format that the SPM Anatomy Toolbox can read. The list of sample coordinates can be read into the Toolbox using the batch cytoarchitectonic coordinate identification tool.
Documentation and Resources
| sample_anatomy_toolbox_demo.m | Export human microarray samples for the SPM Anatomy Toolbox. |
| download_specimen.m | Script for downloading meta information on a Specimen. |
| download_probe.m | Script for downloading meta information on a Probe. |
| download_expression.m | Script for downloading expression values for a set of probes. |
| get_api_path.m | Script that returns the path to the API. |
| transform_samples.m | Transform a set of samples by a given matrix. |
| brodmann.tar.gz | Archived source code. |
| SPM Anatomy Toolbox | Link to the SPM Anatomy Toolbox home page. |